Welcome to the trajalign module distribution homepage!
trajalign is a collection of Python modules that align in space and in time, and average, trajectories of diffraction limited objects, which are recorded with fluorescence microscopy.
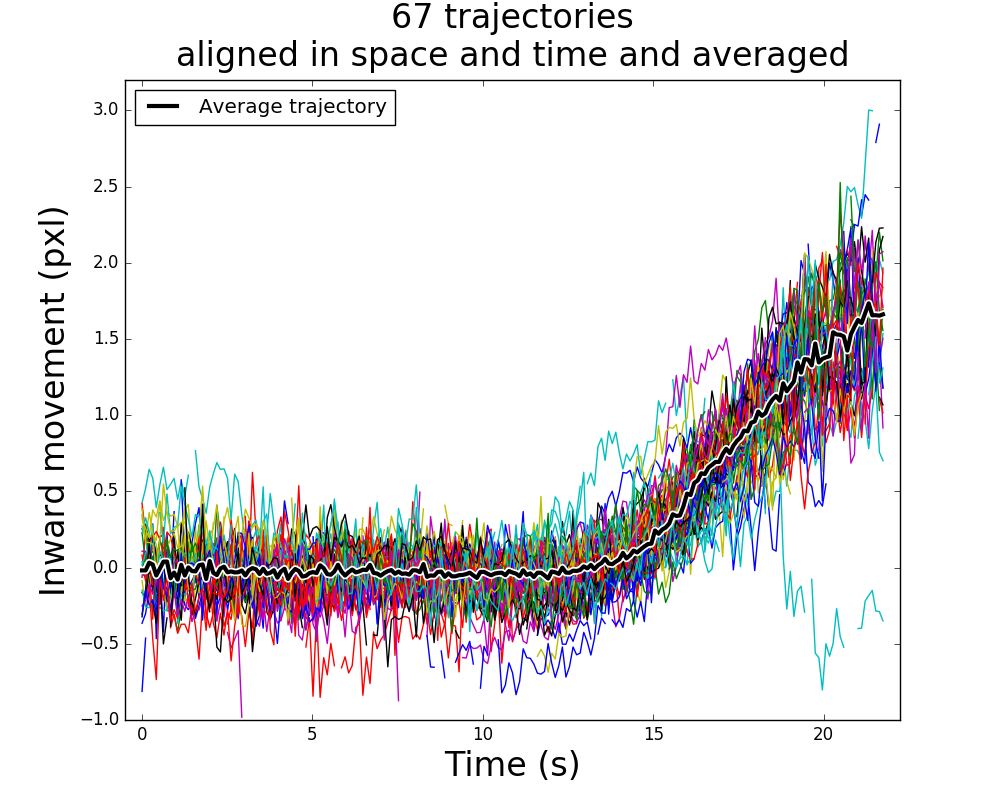 The inward movement of the endocytic coat protein Sla1 during endocytosis. 67 independent trajectories are aligned in space and time and averaged together, with
The inward movement of the endocytic coat protein Sla1 during endocytosis. 67 independent trajectories are aligned in space and time and averaged together, with trajalign module distribution. Generate this plot by following this example.
You can Download the software or visit the github repository. If you already have downloaded trajalign, you might want to check the update history to see whether significant changes have been committed to the repository since your last download.
To install and use the package read the documentation.
If you use this software, please cite: